
The Integrative Human Microbiome Project
Original link:
Https://
The Human Microbiome Project (HMP) was proposed in 2007 and is divided into two phases. The first stage mainly depicts a panoramic view of human microbes for healthy people. The second phase of the Human Microbiome Program, as a continuation of the first phase, explores the role of microbes in health and disease by combining multi-omics research strategies.
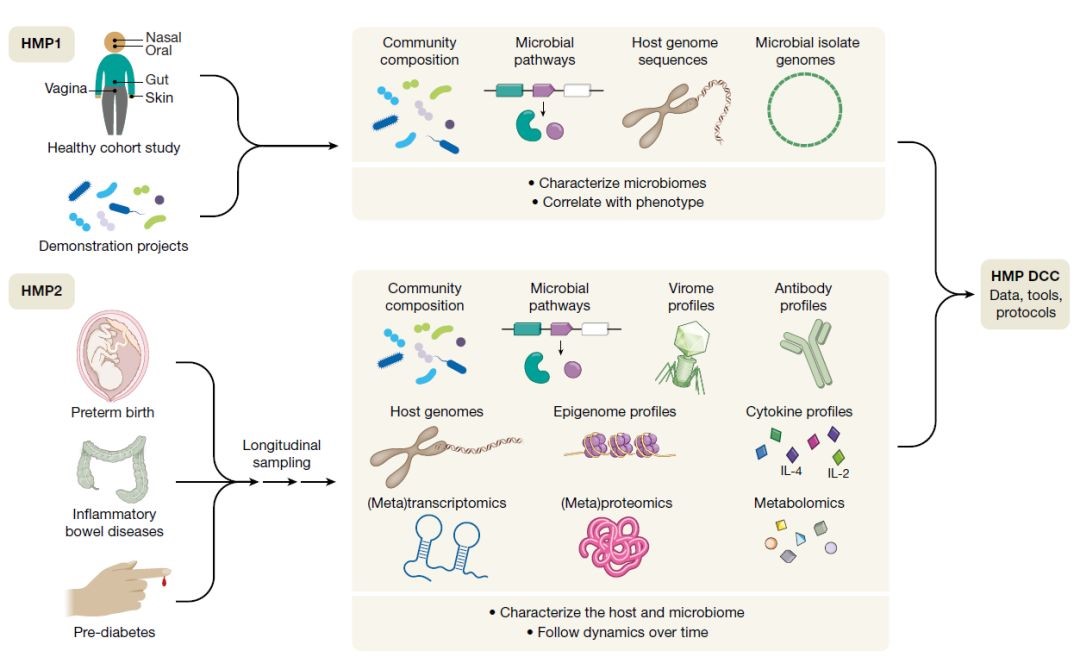
Recently (May 30th), Nature magazine published the completion of the second phase of the Human Microbiome Project, the Integrative Human Microbiome Project (iHMP or HMP2). The program includes three parts of the microbial group and premature labor, microbial and inflammatory bowel disease, microbial group and pre-diabetes, namely the three physiological or pathological states most closely related to the microbiome.
1. "Microbiology and Premature Birth" of the Human Microbiome Program

The vaginal microbiome and preterm birth
Original link:
Https://
Professor Gregory A. Buck of the Virginia Commonwealth University is responsible for research on microbiome and preterm birth in the iHMP project, published in Nature Medicine.
The study collected 12039 samples from 597 pregnant women with different genetic backgrounds, and analyzed 16s diversity, metagenomics , macrotranscripts , cytokines, etc. , and 45 of them were premature pregnant women and 90 full-term pregnant women. (Term birth, TB) conducted a longitudinal analysis and found that:
1. L. crispatus, a strain negatively associated with PTB, enriched in European descent, while BVAB1, a microbe associated with PTB, is enriched in pregnant women of African descent. The findings provide an explanation for the greater risk of premature birth in African descent than in European descent.
2. A proof-of-concept model was constructed from the differential flora of PTB and TB. It was found that BVAB1, Prevotella cluster2, S.amnii and TM7-H1 were able to predict PTB risk. Among them, both BVAB1 and TM7-H1 bacteria have the ability to produce pyruvic acid, acetic acid, 1-lactic acid and propionic acid. It is speculated that the short-chain fatty acids produced by microorganisms may promote inflammation and lead to PTB.
3. The differential flora of PTB and TB is highly correlated with the expression of pro-inflammatory cytokines, suggesting a complex interaction between host and microbes during pregnancy.
Overall, the article provided us with new multi-group big data and helped us to further understand the differences in PTB-related microbial markers in different genetic backgrounds, as well as through microbes, metabolism and immunity. The marker predicts the potential for PTB risk and suggests that this risk assessment should be performed early in pregnancy.
2. "Microbial and inflammatory bowel disease" of the Human Microbiome Program

Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases
Original link:
Https://doi.org/10.1038/s41586-019-1237-9
The team led by Prof. Curtis Huttenhower of the Massachusetts Institute of Technology-Board Institute at Harvard University is responsible for the research on microbiome and inflammatory bowel disease in the iHMP project. The research results are published in Nature.
The study enrolled 132 subjects from 5 academic medical centers. Based on initial endoscopic and histopathological findings, individuals who were not diagnosed with IBD were used as controls, 651 biopsies, 529 blood, and 1785. The stool samples were analyzed by metagenomics, transcriptome, proteome, metabolome and other groups. The results showed that:
1. The author defines a sample that is significantly different from the non-IBD control sample classification as an ecological disorder sample. Compared with non-disordered samples, SCFAs were generally reduced in dysregulated samples, and glycine chenodeoxycholic acid, primary cholate, and glycine and taurine conjugates in CD patients were enriched.
2. The microbial translocation rate of patients with CD and UC is higher than that of non-IBD patients, and the microbial translocation of non-IBD patients mainly occurs in individuals with high abundance of Przetia, while the population shift status of IBD patients reflects early observation. The obligate anaerobic bacteria are relatively reduced and the overgrowth of facultative anaerobic bacteria is often associated with bacterial flora and dysfunction.
3. Differential genes in the ileum (from CD patients) and rectum (from CD and UC patients) and non-IBD patients can directly affect the genes of commensal microorganisms, and these genes can be significantly enriched in immune-related pathways. Especially the IL-17 signaling pathway.
4. The authors found that changes in the abundance of some flora, such as F. prausnitzii, Subdoligranulum-related flora, Escherichia coli and Roseburia, can lead to host metabolic disorders. Some of the key metabolic node products in the associated network, such as acylcarnitine, bile acids, and short-chain fatty acids, propionate, may be dysregulated in the pathological state of IBD.
This study provides the most comprehensive discussion of host and microbial activity in IBD to date. These analyses can be applied to the clinic in future studies, discovering new biomarkers to more accurately predict IBD production and development, and finding new hosts. - Microbial interaction targets provide a breakthrough for relieving or treating IBD.
3. Microbiology and Pre-diabetes in the Human Microbiome Program
Longitudinal multi-omics of host–microbe dynamics in prediabetes
Original link:
Https://doi.org/10.1038/s41586-019-1236-x
Professor Michael Snyder of Stanford University School of Medicine and Professor George Weinstock of the Jackson Genomics Laboratory led the team to conduct research on microbiome and pre-diabetes in the iHMP project. The results were published in Nature.
The study conducted a healthy visit to 106 subjects every 3 months for 4 years. Blood, feces and nasal swab samples were collected from the subjects' health status and illness status, combined with 16S diversity analysis, proteome, Metabolomics, transcriptomes, and genomes, and other omics techniques, found that:
1. The clinical laboratory test data and cytokine individualized differences are the most significant, while the transcripts are not significantly different among the samples; the difference of intestinal microflora between individuals is more significant than that of intestinal microbial predictive genes.
2. RVI and extensive changes in molecular pathways after vaccination, and identification of dysregulated molecular pathways associated with immune responses, metabolism, and neural pathways, while intestinal and nasal microbial homeostasis have also changed.
3. Multi-omics technology can better screen out stress-stimulation events such as viral infection or vaccination from healthy time points.
4. In insulin-sensitive and insulin-resistant subjects, intestinal microbes have different coordination effects with host immunity and metabolism.
5. By examining the multi-omics test data during the T2D development of the subject, the authors identified hundreds of molecules that changed prior to the diagnosis of the disease, and these molecules are likely to be potentially associated with the T2D mechanism of the subject. Correlation.
This study used multi-omics sequencing to reveal the differences in biological pathways and physiological responses between individuals with glucose metabolism and healthy individuals under healthy conditions and disease, and to further study health status, pre-diabetes status and T2D status. The difference between the intrinsic link provides an open source database. Subsequent research will help to understand how multi-omics factors affect human health at the group and individual levels and determine how they mutate early in the disease to prevent disease.
Fourth, the conclusion
The joint results of the National Institutes of Health HMP project and many other projects show that microbial communities are an integral part of human biology and play an important role in health and well-being. Along with the end of HMP, it also brings more questions to answer: many immune and biochemical reactions seem to be related to specific strains, which is unique to one or several individual individuals, but it is not clear yet. The strain is the cause for the disease. The human-related microbiome has evolved from infectious diseases and gastrointestinal diseases to a wider range of areas in the last decade, including metabolism, tumors, maternal and child health, and functions of the central nervous system . With the end of the NIH HMP project, it will showcase the future development of new research and new technologies. We also look forward to getting more new discoveries from the resources of the project.
White Sesame Oil,Ground Sesame Seed Oil,White Sesame Seed Oil,A Grade Sesame Oil
Chinese Seasoning (Shandong) Trading Co.,Ltd , https://www.zt-trading.com