First, the screening of large-scale gene function
Despite significant advances in sequencing and genome editing techniques, the analysis of complex genotype-phenotype relationships remains a major obstacle to quantitative genetics. As a powerful gene editing tool, the CRISPR-Cas9 system can generate frameshift mutations in the exon region of protein-coding genes and completely destroy protein expression and function. This property is widely used in large-scale functional screening of genes. It will also greatly accelerate the assessment of the functional impact of genetic variation.
1. Using C RISPRa to construct an unbiased drug-resistant Ln cRNA screening platform
A study published in the April 2018 issue of the Cell Journal developed a groundbreaking approach to identify and determine the functional role of lncRNA in the development of resistance to chemotherapeutic drugs in acute myeloid leukemia (AML). This technology screens both coding and non-coding genes that affect therapeutic responses by combining information from publicly available pharmacological databases with CRISPR technology. As a genome-wide screening platform, it does not favor coding genes, nor is it biased towards non-coding genes, and can screen for new therapeutic targets [1 ] .
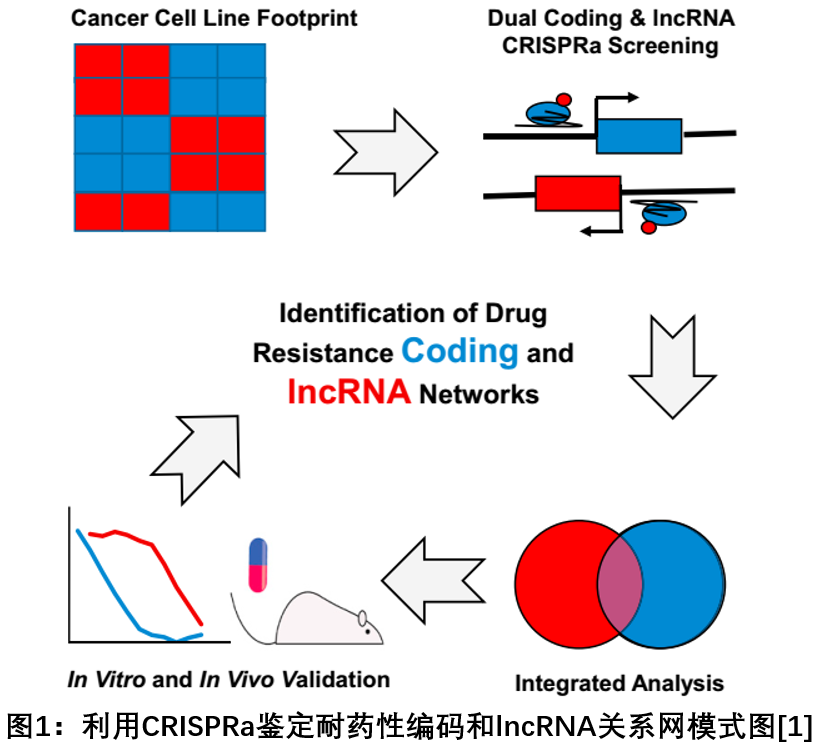
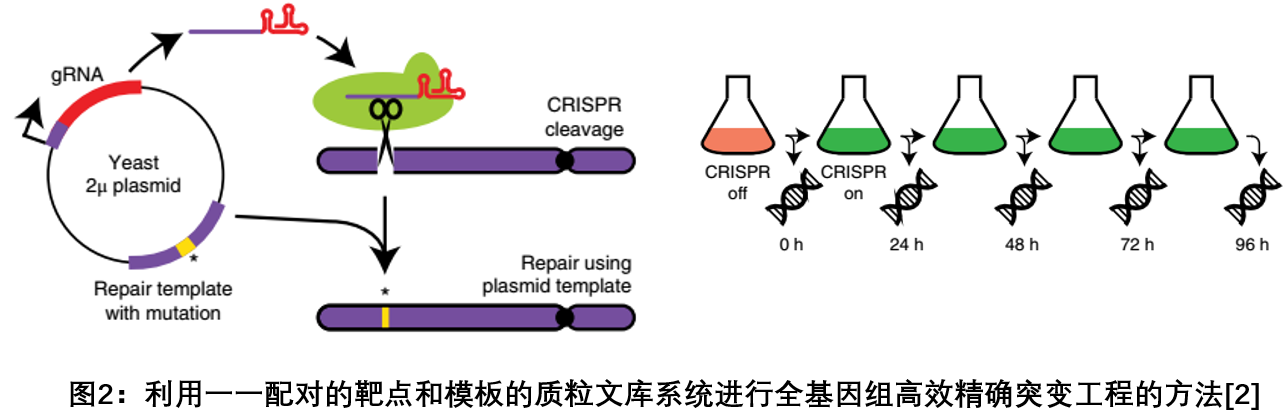
2. Use a one-to-one paired target-template strategy to achieve genome-wide efficient and accurate mutations
Understanding the functional effects of DNA mutants is critical for basic biology, evolutionary biology, and medical genetics; although CRISPR-Cas9 technology can achieve the potential for multi-gene induced mutations in thousands of cells in vivo, high throughput The specificity of sequencing these mutants remains a technical bottleneck. In April 2018, the Nat Genet journal published a study that solved the current barrier of tens of thousands of genome-edited results in high-throughput sequencing by transforming the CRISPR-Cas9 system and developed a CRISPR-based genome-wide efficiency and precision. The method of mutation engineering. As shown, 10,000 pairs of plasmid libraries encoding gRNA targeting sequences and their corresponding cis-repair templates were constructed to achieve one-to-one pairing of targets and templates, and libraries were delivered to yeast cells to monitor the effects of gene mutations. The efficiency is as high as 95%. This strategy can efficiently and accurately track the effects of a large number of gene mutations on cell function, providing a powerful analytical tool for studying gene function [2 ] .
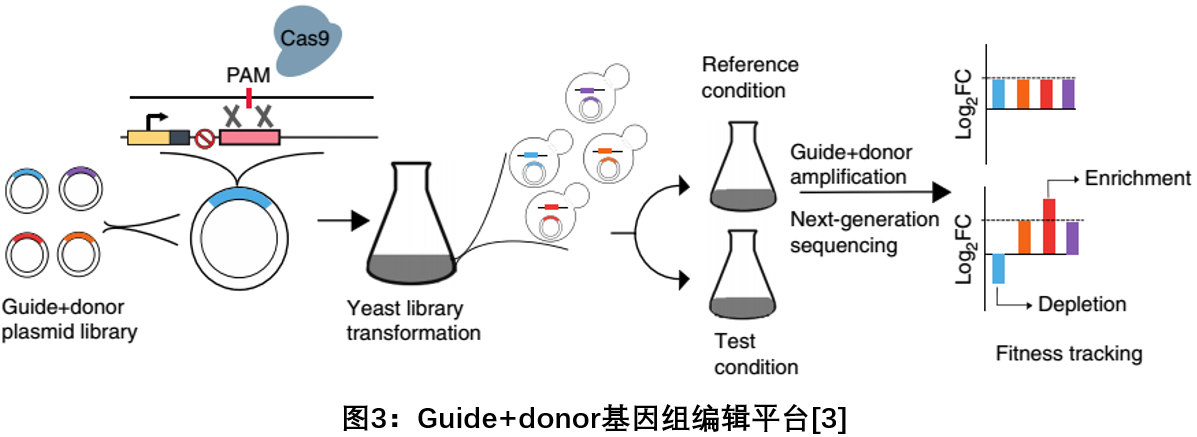
3. Guide + donor: C RISPR-C 9 using a high throughput functional in yeast Construction and analysis of DNA sequence variation in the library as
In May 2018, Harvard Gene Editor Daniel Church research team published a report in Nature Biotechnology, introducing a Cas9-based method that efficiently (80-100%) produces specific genetic variants (deletions, substitutions, and insertions) in yeast. Library, and the overall approach to tracking its adaptability. The guide+donor method was used to accurately eliminate 315 genes in the yeast genome, and to evaluate and identify genes that play an important role in cell adaptive survival. The new method can not only accurately perform functional genomic research in yeast in a high-throughput manner, but also deepen the function of low-frequency gene mutations and non-coding sequences, opening a new door for gene function analysis [3 ] .
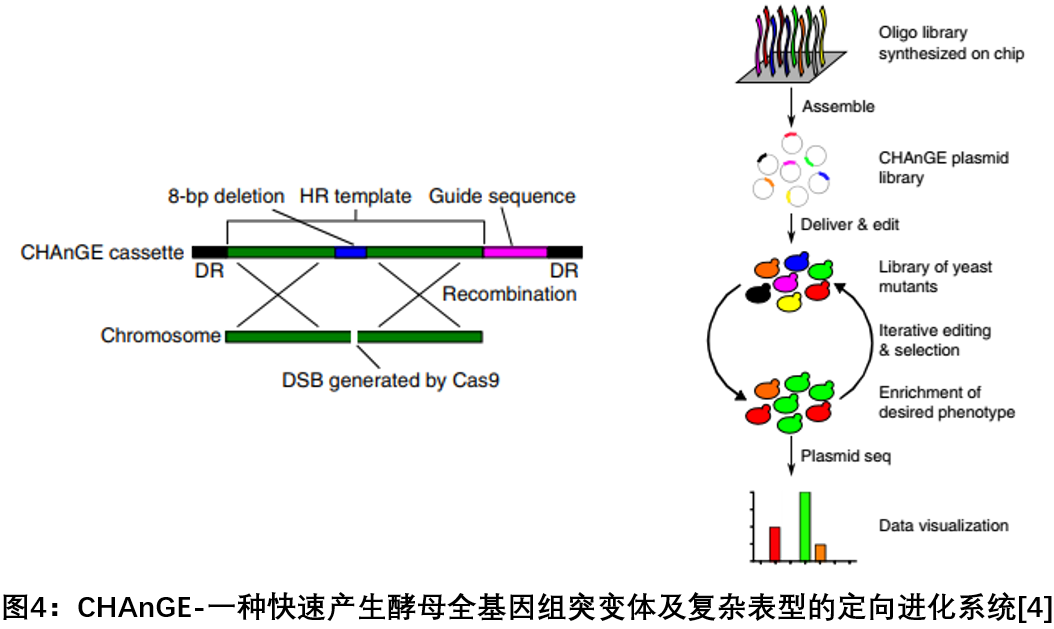
4. CHAnGE: a large-scale genome screening method based on C RISPR-C as 9 and homologous directed repair
In July 2018, the research team of the University of Illinois, Zhao Huimin, published a report on Nature Biotechnology, which developed a large-scale genome screening method based on CRISPR-Cas9 and homologous directed repair. CHAnGE can rapidly generate thousands of species across the genome. Accurate yeast single nucleotide mutants, tracking the effects of gene mutations on cells with a mutation efficiency of over 98%. This study verified the feasibility of this method for single-nucleotide resolution genome editing by creating a genome-wide gene mutation set, and solved the problem of previous method editing efficiency limitations. This method enables rapid genome-wide engineering of Saccharomyces cerevisiae and precise and traceable gene mutations [4 ] .
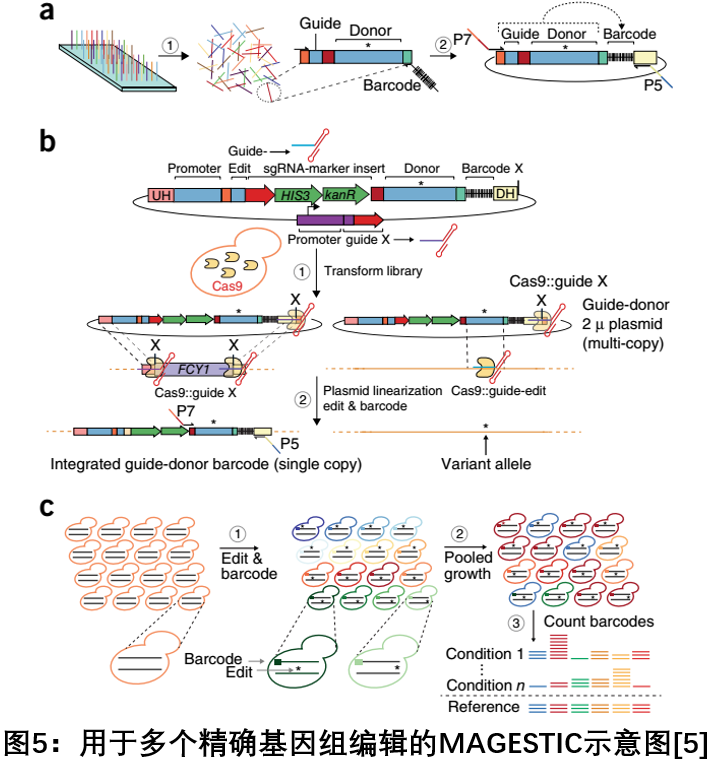
5. MAGESTI C : A Multiple Exact Genome Editing Technique for Tracking Genomic Barcodes in Yeast
In July 2018, the Steinmetz LM team at Stanford University in the United States published a report on Nature Biotechnology to develop a method for multiple precise genome editing in Saccharomyces cerevisiae based on Cas9 technology using a short, traceable integrated cellular barcode (MAGESTIC). . MAGESTIC uses a plasmid system of synthetic array gRNA-donor oligonucleotides for high-throughput editing with genomic barcode integration to prevent loss of plasmid barcodes and maintain a stable phenotype. The use of the LexA-FKH1P fusion system to actively recruit donor DNA to DNA breaks can increase the efficiency of homologous recombination. And it has a greater advantage than the way that donor merges into Cas9. On the one hand, it can recruit more copies of donors, and it does not depend on the continuous association between Cas9 and DSBs. This technique overcomes several limitations of currently available methods, namely the instability of plasmid barcodes, and the inability to distinguish between Oligo synthesis and PCR or sequencing errors in targeting RNA and donor templates during phenotypic analysis. MAGESTIC technology will be widely used to reveal the genetic basis of yeast phenotype [5 ] .
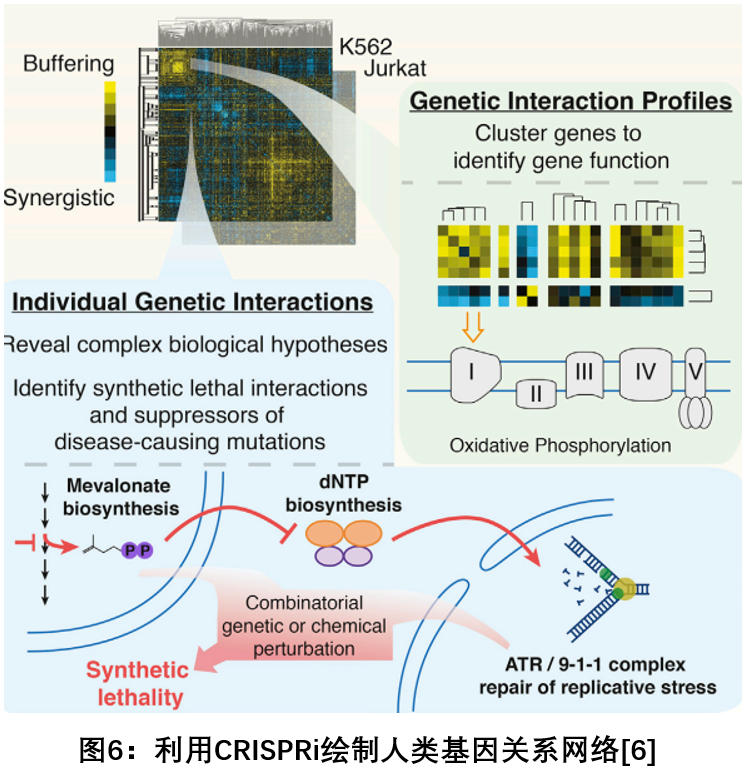
6. Rapid mapping of human genes based on C RISPRi 's high-throughput technology
In July 2018, Cell published a research study by the University of California, San Francisco, and developed a CRISPR-based high-throughput technology to rapidly map the function of nearly 500 genes in human cells, many of which were previously Has never been studied in detail. The genes that humans have studied so far are less than 10%, and we still know nothing about the remaining gene functions. The methods traditionally required to test gene function are both expensive and time consuming. Developing a rapid and comprehensive determination of these unresearched gene functions will help scientists gain a broader understanding of biology. The study used a technique called genetic interaction mapping to establish a comprehensive analysis of gene function in humans. Using a tool called CRISPR Inhibition (CRISPRi), you can reduce gene activity without editing the DNA itself, systematically shut down the paired genes in a single cell and measure how the cells react, interpreting the two genes The relationship between genes that can quickly classify unknown functions. Using this system, the researchers identified new genes involved in cellular energy production and explained why some cholesterol drugs can be used to treat osteoporosis and the long-term mystery of related drugs. The ultimate goal of the study is to plan to map the entire genome of the entire human genome [6 ] .
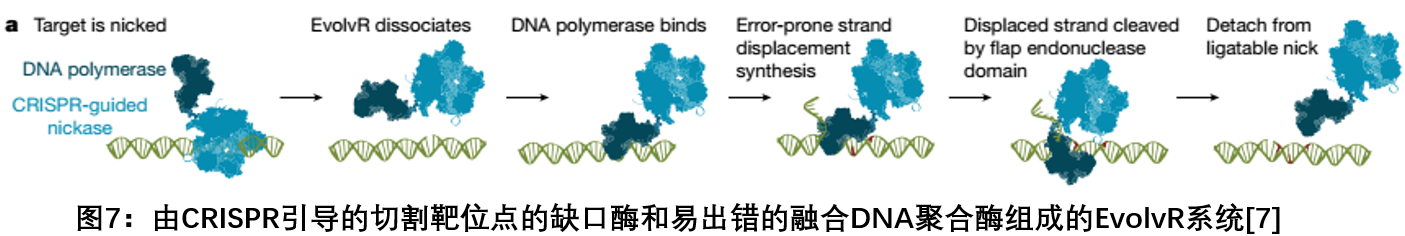
7. EvolvR: A platform that combines CRISPR and DNA polymerase to promote the evolution of specific genes in cells
In August 2018, the Institute of Innovative Genomics at the University of California at Berkeley published a report on Nature, proposing a revolutionary new approach that harnesses the power of natural evolution—a platform that promotes the evolution of specific genes within cells. They named the new system "EvolvR", which fuses Cas9 with a DNA polymerase, using a nicking-Cas9 to cut only one of the two DNA strands, and the special incision is filled by the polymerase. . The process of DNA polymerase complementation produces errors that result in various types of mutations. Therefore, this error in polymerase can bring more genetic diversity, using EvolvR to artificially create random mutations, creating millions of different combinations of sequences, and discovering many mutant species that are needed by humans. This system can even achieve thousands of different genetic diversifications in an experiment, creating new functions, not just opening and closing genes [7] .
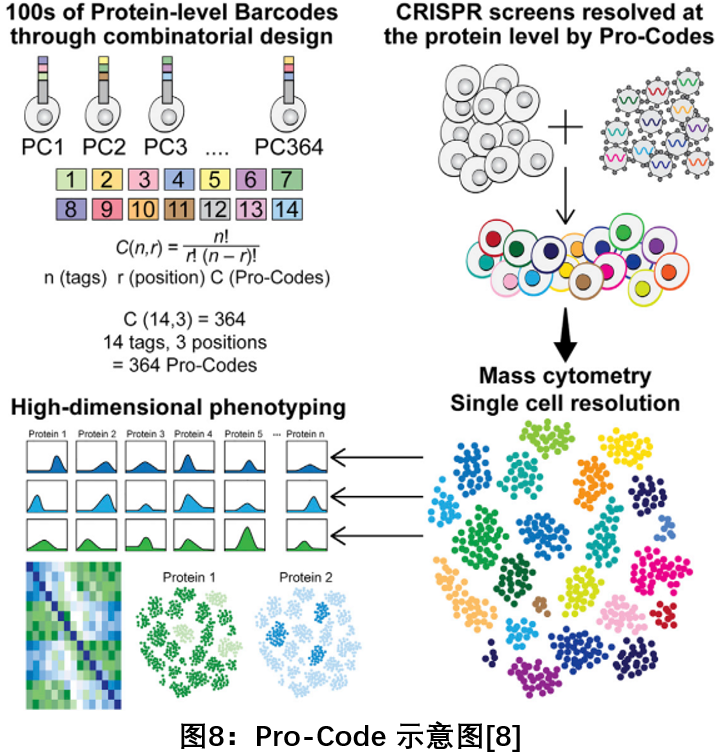
8. Pro – Codes: A system for high-resolution analysis of hundreds of gene functions using protein barcode technology
In October 2018, researchers from Mount Sinai Hospital published a paper in the Cell Journal, developing a protein-level barcode system (Pro-Codes) that uses a combination of linear epitopes to create higher-magnification protein barcodes. Hundreds of gene functions can be analyzed simultaneously with resolutions up to single cell levels. Using synthetic protein "epitopes" to tag and track different CRISPRs, Pro-codes can simultaneously encode hundreds of CRISPRs to knock out large numbers of genes and analyze them by mass spectrometry. Currently, genetic screening techniques using DNA as a barcode can only achieve a limited phenotype and a rough cell resolution. The new Pro-Codes technology enables high-dimensional protein-level phenotypic analysis of 100 genes simultaneously at single-cell resolution, allowing a more comprehensive description of the biological effects of individual genes [8] .
9. screened functional Lnc RNA genome a new policy by C RISPR-C as 9 targeted cleavage site
In November 2018, in order to make up for the high-throughput functional screening of lncRNA for large-scale genomic deletion, CRISPRi and CRISPRa, the deficiency in efficiency, quality (false positive, false negative), Peking University Wei Wensheng research group in Nature Biotechnology magazine has published a new screening strategy to generate exon deletions or intron retention of genes in a high-throughput manner by constructing a novel CRISPR library that specifically targets the splice sites of the gene. Using this strategy, efficient screening of lncRNA function at the genome-wide level was achieved, and 230 lncRNAs were screened and found to be involved in cell survival or proliferation. Further analysis showed that lncRNA function has significant heterogeneity in different cell types. The establishment of this new high-throughput technology platform is the first to achieve a high-throughput screening based on complete knockout of long-chain non-coding RNAs at the genome-wide level. This methodology will provide an effective tool for systematic discovery and analysis of lncRNA functions. [9 ] .
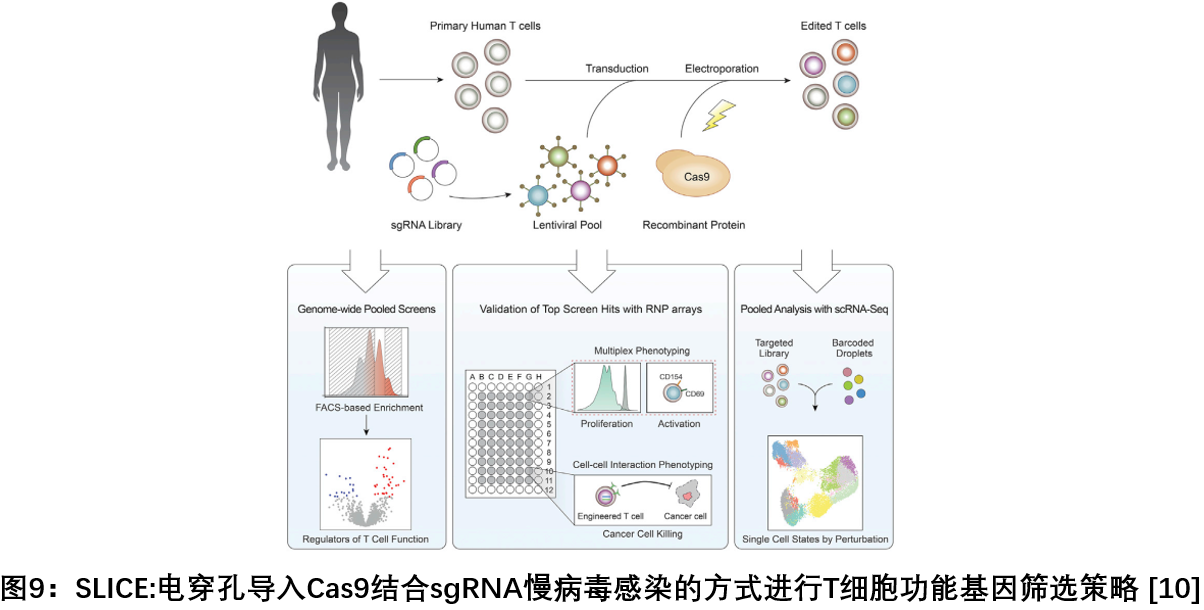
10. SLICE: A system for single-guide RNA lentivirus infection using electroporation of Cas 9 protein
In November 2018, researchers from the University of California, San Francisco, published a paper in Cell, proposing a CRISPR-based screening tool, SLICE, a system for single-pass RNA lentivirus infection using Cas9 protein electroporation. Traditional CRISPR Screening Technique Based on sgRNA Lentiviral Library Due to the long time that the lentiviral vector encodes the Cas9 protein, it is restricted for primary T cells with limited culture time in vitro, and the SLICE system will be electroporated by sgRNA lentiviral library and Cas9 protein. In a combined manner, it is possible to rapidly introduce into the patient's primary T cells and evaluate the function of each gene. This new method can provide a strong basis for the transformation of immune cells against cancer and the guidance of other diseases [10 ] .
Second, D NA labeling and cell lineage tracing
The focus of developmental biology includes the diversity of cell types that make up an organ or organism and the developmental lineage history of these cells. These aspects are usually studied separately as a single direction, but four recent papers have reported a single-cell RNA sequencing (scRNA-seq) technique combined with CRISPR-based lineage tracing to simultaneously dissect transcriptome cell phenotypes. And the method of pedigree history. In recent years, the CRISPR-Cas9 gene editing system has been proven to be useful for cell lineage tracing [11 ] . In a classical protocol, barcode arrays are designed into the genome of an organism; in the early stages of embryonic development, embryos are co-injected with Cas9 and targeting RNA (gRNA) to target a barcode array. During the editing of the CRISPR, the barcode is cumulatively cut and repaired in a variable manner in different cells. Subsequent analysis of the barcode sequence at a specific development time point can infer the lineage relationship between cells from the existing Cas9-induced mutation types.
1. Three landmark studies used the cell type information provided by scRNA-seq to study the development of zebrafish, demonstrating the feasibility of the CRISPR system in cell lineage tracing
Alemany et al. developed a method called ScarTrace , which is a transgenic genomic cluster that targets eight tandem copies of the GFP gene after the Cas9 protein or RNA is injected with the gRNA targeting GFP. For scRNA-seq analysis of differentiated tissues, the team used a sorting and automated assisted transcriptome sequencing (SORT-seq) protocol. Although the GFP sequence edited by Cas9 ('scarlet') is usually expressed in cells, it can be read from the transcriptome, but SORT-seq binds to the nested PCR step to read scar sequences from the remaining genomic DNA in a single cell. , thereby minimizing the loss of lineage information of cells lacking GFP expression. The researchers analyzed the clonal history of various tissues - including hematopoietic systems, adult brains and eyes, and regenerative fins - to provide various developmental information, such as the number and type of progenitor cells that are precursors of specific differentiated cell subsets [12 ] .
l Spanjaard et al. reported a pedigree tracking method similar in concept to the former, editing ubiquitous sequences (LINNAEUS) by nuclease activation, allowing one to determine cell types and cell lineages. The method involves embryo injection of Cas9 protein and gRNA targeting 16-32 integration sites of the red fluorescent protein (RFP) gene throughout the genome. The basic principle of using a genome-dispersed target sequence is to avoid deletions between nearby array copies, which may delete copies of large amounts of information between them. LINNAEUS is suitable for zebrafish larvae, as well as adult heart, liver, pancreas and brain. The team performed a scRNA-seq using droplet-based microfluids, and the mutated RFP sequences were read from the transcriptome and divided into a library for transcriptome range analysis and a library targeted to analyze RFP sequences. In addition to the experimental process, Spanjaard et al. highlighted the challenges of bioinformatics and the need for custom analysis methods, as the standard phylogenetic algorithms derived from species evolution studies are not optimal for cell lineage reconstruction, mainly due to the large number of Single cell comparisons and pedigree barcodes (tens of thousands) cannot be fully detected [13 ] .
In the third study, Raj et al. focused on how to overcome the limitations of embryonic injection of Cas9 only during the first few hours of development. Therefore, in addition to embryonic injection of Cas9 protein and four gRNAs targeting 1-4 sites targeting RFP barcodes, zebrafish also transgene encodes Cas9 and five additional gRNAs targeting the 5-9 sites of RFP barcodes, these gRNAs The stage of late development can be monitored by heat induction at a specific time. This method is called scGESTALT (a single-cell-derived technique based on the previously reported GESTALT tracer method), and scRNA-seq is similar to Spanjaard et al., using droplet-based microfluidics and targeted enrichment of transcriptomes. To sequence the barcode. Raj et al.'s analysis focused on the brain, identifying many lineage relationships between neural cell types and possible gene expression relationships during developmental transitions [14 ] . These powerful cell tracer techniques can be used to study a variety of normal and pathological developmental phenomena and can be used in other model organisms.
2. "founder mouse (founder mouse)" is a cell lineage tracing CRISPR system in mammalian cells may be created
These studies have demonstrated the feasibility of applying the CRISPR-Cas9 gene editing system to cell lineage tracing in cells and lower vertebrates. However, it has not yet been validated in mice, a model organism that is more relevant to human health in many ways, such as development. The mouse develops longer and differentiates into many lineages throughout its development, thus requiring the continued generation of highly diverse barcodes, minimizing unnecessary rewrite events to maximize the success of recording important events. probability.
In a new study, the George Church team from Harvard University constructed a small "initiating mouse" containing multiple independent barcode site-dispersed arrays (hgRNAs) to create a small mouse and a small Cas9 protein. In mouse hybridization, progeny mice produce a unique mutation in each lineage because hgRNA randomly accumulates mutations throughout pregnancy, and does not delete early mutations, allowing related cells to have more similar mutation profiles or barcodes. In developmental bar code mice, the activity profiles and mutant alleles of each hgRNA are extensively characterized, and the bottom line of the lineage tree is reconstructed early in the early development, starting from the first branch of the root and tracking the cells. The original pedigree. This study provides a viable universal platform system for in vivo barcode and lineage tracking in mammalian models [15 ] .
Third, the disease diagnosis technology based on CRISPR-Cas system
Rapid molecular diagnostic techniques are very useful in many areas, including public health, environmental testing, and criminal investigation. The in vitro diagnostic technology developed based on CRISPR was selected into the world's top ten scientific and technological advances in 2018. The CRISPR-Cas system has now been shown to specifically recognize and cleave DNA and RNA, suggesting the potential use of this system as a universal nucleic acid recognition tool. Due to the specificity of recognition between the target sequence and the CRISPR-Cas system, the CRISPR-Cas system has been used to detect point mutations or single nucleotide variants. This technology detects pathogens during infection by analyzing the DNA or RNA of the pathogen. The following is a list of major breakthroughs in molecular diagnostics for the CRISPR-Cas system this year (shown below).
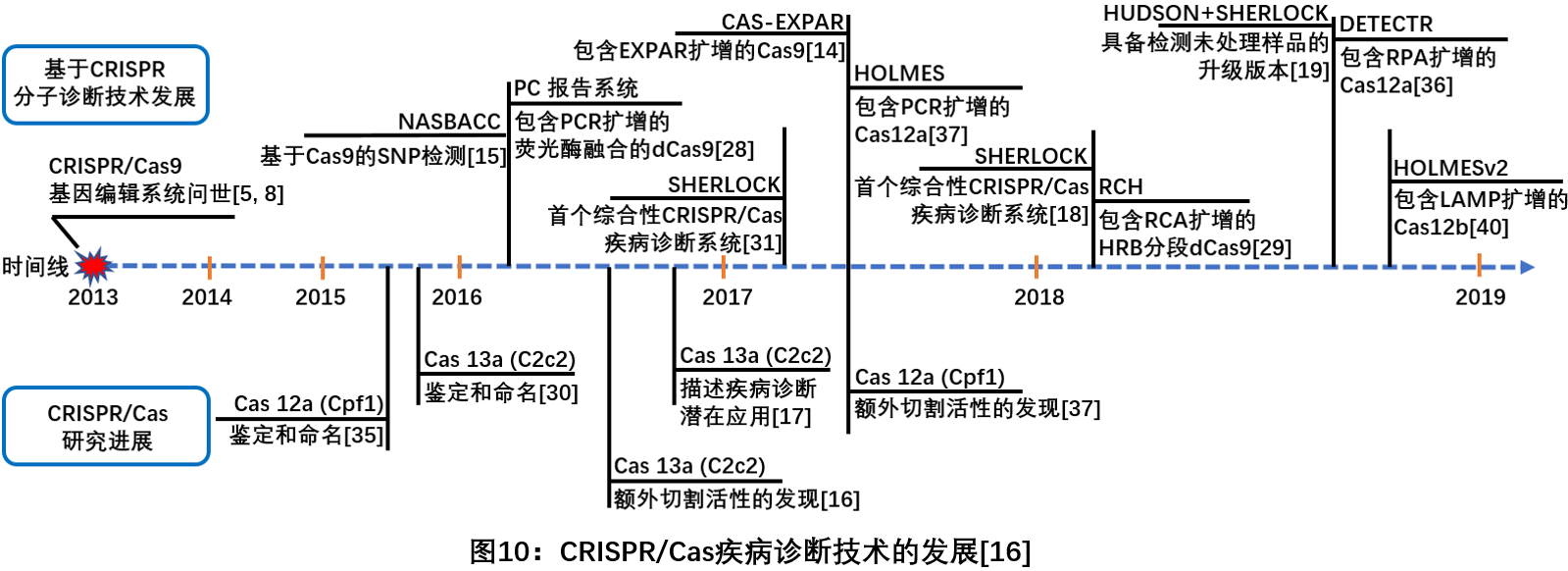
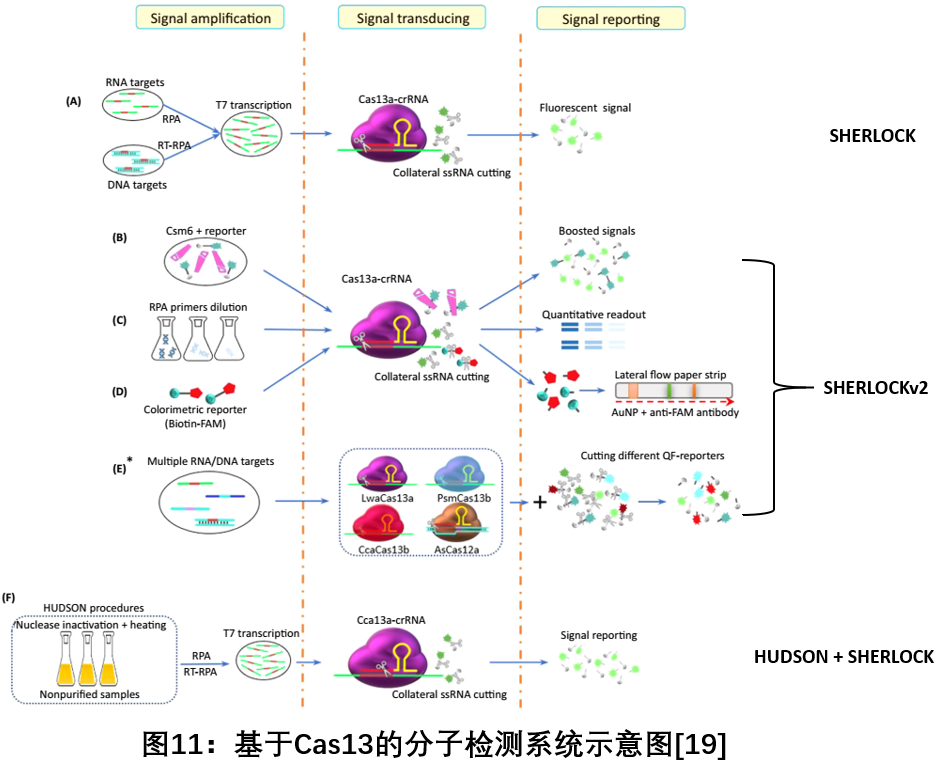
1. Based on the molecular detection system Cas13
 Since 2017, Zhang Feng team used Cas13a (formerly known as C2c2) to combine the "collateral effect" of hybrid ribonuclease activity with target isothermal amplification in target recognition, and established a CRISPR-based diagnostic method ( CRISPR-Dx). A rapid DNA or RNA detection technology with the same sensitivity and single base mismatch specificity - SHERLOCK (Specific High-sensitivity Enzymatic Reporter unLOCKing) diagnostic platform, can be used to detect Zika virus and dengue virus, bacteria Isolates, antibiotic resistance genes, human DNA genotypes, and cancer mutations [17 ] . Soon after, the team optimized the SHERLOCK diagnostic platform and successfully developed the SHERLOCKv2 system, which integrates four orthogonal CRISPR-Cas enzymes and is the only detection system that can simultaneously detect multiple target sites. To highlight the potential of its multi-directional, portable, rapid and quantitative nucleic acid detection platform [18 ] . At the same time, they have further developed the HUDSON (heating unextracted diagnostic samples to obliterate nucleases) system, which complements SHERLOCK and is used to detect viruses directly from body fluids. It can be directly sampled from patients in less than 2 hours without any instrument. The detection in the middle greatly improves the convenience of molecular detection [19 ] .
Since 2017, Zhang Feng team used Cas13a (formerly known as C2c2) to combine the "collateral effect" of hybrid ribonuclease activity with target isothermal amplification in target recognition, and established a CRISPR-based diagnostic method ( CRISPR-Dx). A rapid DNA or RNA detection technology with the same sensitivity and single base mismatch specificity - SHERLOCK (Specific High-sensitivity Enzymatic Reporter unLOCKing) diagnostic platform, can be used to detect Zika virus and dengue virus, bacteria Isolates, antibiotic resistance genes, human DNA genotypes, and cancer mutations [17 ] . Soon after, the team optimized the SHERLOCK diagnostic platform and successfully developed the SHERLOCKv2 system, which integrates four orthogonal CRISPR-Cas enzymes and is the only detection system that can simultaneously detect multiple target sites. To highlight the potential of its multi-directional, portable, rapid and quantitative nucleic acid detection platform [18 ] . At the same time, they have further developed the HUDSON (heating unextracted diagnostic samples to obliterate nucleases) system, which complements SHERLOCK and is used to detect viruses directly from body fluids. It can be directly sampled from patients in less than 2 hours without any instrument. The detection in the middle greatly improves the convenience of molecular detection [19 ] .
2. Based on the molecular detection system Cas12
The Jennifer A. Doudna team also achieved sensitive and accurate DNA detection by combining Cas12a (CpfI) with a molecular signal gun. This method is called DETECTR (DNA Endonuclease Targeted CRISPR Trans Reporter). The basis of this approach is the discovery that Cas12a has the ability to cleave all single-stranded DNA in the vicinity of a DNA target, and the developer uses a fluorescent molecule to link through a single-stranded DNA to another inhibitory molecule that inhibits the luminescence of this fluorescent molecule. Together. After the single-stranded DNA is cleaved by Cas12a, the ruthenium will be removed, allowing this fluorescent molecule to illuminate. Using this technique, they successfully performed a correct judgment on carcinogenic HPV patient samples [ 20 ] . At the same time, my King Jin team published a research paper titled CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA in the March 12 issue of Cell Research, as published by the previous Jennifer Doudna team. The results of the study were similar, systematically studied the cleavage properties of Cas12a for target ssDNA and non-target ssDNA [ 21 ] .
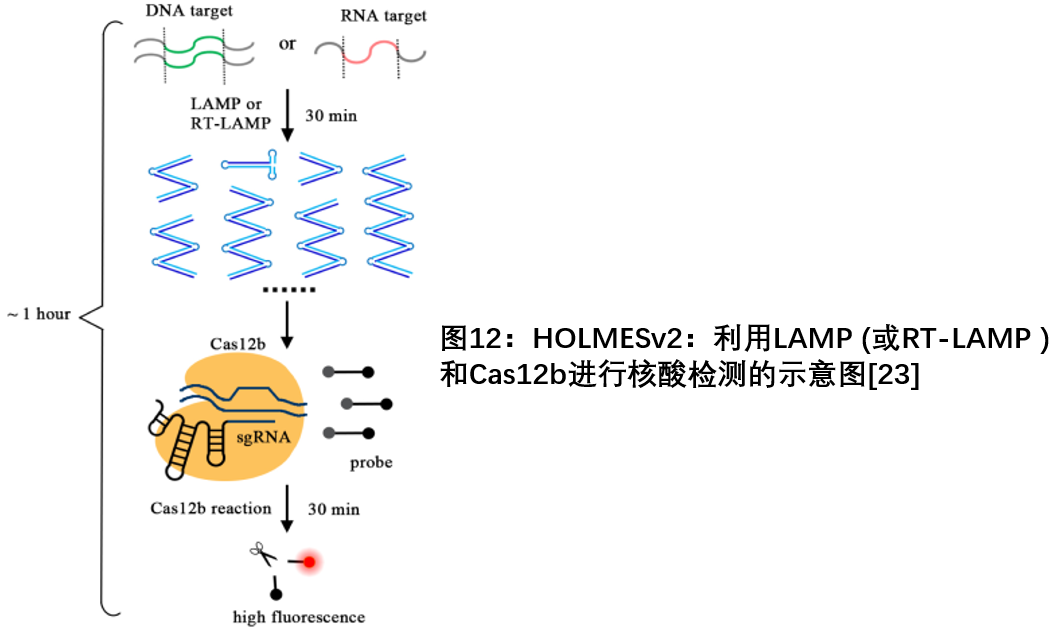
In addition, the Shanghai Tolo Harbour research team developed a highly efficient, sensitive and specific platform for detecting DNA, RNA viruses and human SNPs based on Cas12a with trans-cleavage activity on side-chain single-stranded DNA in 2017. -HOur Low-cost Multipurpose highly Efficient System) [ 22 ] . Then at the end of 2018, the team combined the thermophilic Cas12b with ssDNA trans-cleavage activity with LAMP-mediated isothermal amplification (LAMP) to create an improved version of HOLMESv2. In HOLMESv2, LAMP amplification and Cas12b trans-cleavage can be integrated into a constant temperature one-step system, thus bringing great convenience to nucleic acid detection. They also simplified the RNA detection program in HOLMESv2, using RNA-dependent DNA polymerase for amplification, thus omitting additional reverse transcription steps, which is by far the simplest CRISPR biosensing system for RNA detection [ 23 ] .
summary:
In summary, we introduced the progress of CRISPR/Cas9 in 2018 and the application of many aspects. It can be seen that basic scientists have made great progress in understanding and manipulating biology using CRISPR-Cas9 technology. In clinical, we can Expect new treatments for genetic diseases (such as using Cas9 gene editing or CRISPRi/a) and new diagnostic techniques. We are able to use nature's technology toolbox to evolve and transform more efficient and secure genetic editing tools for us. The new CRISPR system may be hidden in the genome of the organisms around us. In the future, there may be more CRISPR derivatives with new mechanisms and functions, which will provide powerful breakthrough technologies for human beings.
references:
[1] Bester, Assaf C., et al. "An Integrated Genome-wide CRISPRa Approach to Functionalize lncRNAs in Drug Resistance." Cell173.3(2018): 649-664.e20.
[2] Sadhu, MJ, et al. "Highly parallel genome variant engineering with CRISPR-Cas9. " Nature Genetics 50.4 (2017): 510.
[3] Guo, Xiaoge, et al. "High-throughput creation and functional profiling of DNA sequence variant libraries using CRISPR–Cas9 in yeast." Nature Biotechnology (2018).
[4] Bao, Zehua, et al. "Genome-scale engineering of Saccharomyces cerevisiae with single-nucleotide precision." Nature Biotechnology (2018).
[5] Roy, KR, et al. "Multiplexed precision genome editing with trackable genomic barcodes in yeast." Nature Biotechnology (2018).
[6] Horlbeck, Max A., et al. "Mapping the Genetic Landscape of Human Cells." Cell (2018): Â S0092867418307359-.
[7] Halperin, Shakked O., et al. "CRISPR-guided DNA polymerases enable diversification of all nucleotides in a tunable window." Nature (2018).
[8] Wroblewska, A, et al. "Protein Barcodes Enable High-Dimensional Single-Cell CRISPR Screens." Cell175.4(2018):1141.
[9] Liu, Ying, et al. "Genome-wide screening for functional long noncoding RNAs in human cells by Cas9 targeting of splice sites." Nature Biotechnology (2018).
[10] Shifrut E, et al. Genome-wide CRISPR Screens in Primary Human T Cells Reveal Key Regulators of Immune Function. Cell. 2018 Dec 13;175(7): 1958-1971.e15.
[11] Kalhor, Reza, P. Mali, and GM Church. "Rapidly evolving homing CRISPR barcodes." Nature Methods 14.2(2016):195-200.
[12]  Alemany, A. et al. Whole-organism clone tracing using single-cell sequencing. Nature 556, 108–112 (2018)
[13] Spanjaard, Bastiaan, et al. "Simultaneous lineage tracing and cell-type identification using CRISPR–Cas9-induced genetic scars." Nature Biotechnology (2018).
[14] Raj, Bushra, et al. "Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain." Nature Biotechnology (2018).
[15] Reza, Kalhor, et al. "Developmental barcoding of whole mouse via homing  CRISPR." Science (2018):  Eaat9804-.
[16] YiLi et al., CRISPR/Cas Systems towards Next-Generation Biosensing . Trends Biotechnol (201 8 ).
[17] Gootenberg, Jonathan S., et al. "Nucleic acid detection with CRISPR-Cas13a/C2c2." Science 356. Â 6336 Â (2017): 438-442.
[18] Myhrvold, Cameron, et al. "Field-deployable viral diagnostics using CRISPR-Cas13." Science 360.6387 (2018): 444-448.
[19] Gootenberg, Jonathan S., et al. "Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6." Science 360.6387 (2018): eaaq0179.
[20] Chen, Janice S., et al. "CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity." Science (2018): eaar6245.
[21] Li, Shi Yuan, et al. "CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA." Cell Research  (2018).
[22] Cheng, QX et al. Shanghai Tolo Biotech Co., Ltd. An application of a Cas protein, and a method and kit for detecting a target nucleic acid molecule, (2 017 ) CN107488710A
[23] L. Li, S. Li, J. Wang, CRISPR-Cas12b-assisted nucleic acid detection platform. bioRxiv, (2018).
As a professional supplier of medical apparatuses and instruments, Suzhou Medtion International Trade Co., Ltd. is located in Zhangjiagang City, Jiangsu Province. We are wholly owned subsidiary of Jiangsu Redleaf Medcial Equipment Co., Ltd who is a professional Medical equipment producer for 30 years. We specialize in developing and producing stretchers,anti-coronavirus products, first-aid product, Funeral Products, ward products, Diagnosis Device, emergency rescue product and so on. We supply one stop purchasing of Medical Equipment for all overseas customers. Our products can be made in accordance with customers' requirements. With advanced technology, scientific production equipment, reliable product's quality, conform contract, stable credit and excellent service, we have got the production aptitude and passed all quality authentications successfully. We warmly welcome you to cooperate with us.
Operation Room Equipment,Medical Binocular Magnifier,Operation Bed,Operation Instrument
Medton Medical , https://www.medton.cn